Fixing Issues Related to Data (Loading)#
Learning how to deal with certain issues that can arise when using new experimental data.
When experimental data is loaded into a reconstruction software like PtyPy, it is important that the geometry of the experiment is modelled correctly. When populating the parameter tree for a new data set, there are few things that can easily go wrong. The most common issue is that at least one of photon energy, detector distance or detector pixel size has been wrongly defined in the parameter tree. As part of the model initialisation in PtyPy, the reconstructed pixel size is being calculated based on these three physical quantities which might give the following error
RuntimeError: Arrays larger than 50M not supported. You requested 87706.44M pixels.
if they have not been provide correctly.
Note
Since PtyPy 0.8.1 the megapixel limit has been increased to 100M therefore you might run out of memory before seeing this RuntimeError.
Another common issue is that we often don’t know exactly the coordinate system of the detector and the scanning stage that was used during data collection, and even if we know it might not match the default coordinate system used in PtyPy. Luckily, there are only a few (8 to be precise) possible configurations and the PtyScan base class has a parameter called orientation that makes it very simple to do a brute-force search for the correct setup.
Data file |
Type |
Download |
Courtesy of |
Reference |
|---|---|---|---|---|
dls_i08_nanogold_issue.h5 |
Raw data |
Majid Kazemian |
Based on the small I08-1 data set that was used in the tutorial on Loading HDF5 Data we have a created a new file named "dls_i08_nanogold_issue.h5" for which the coordinate system has been modified sligthly. The task for this tutorial is to figure out the correct orientation based on the data that has been provided. As a starting point, we set it to zero.
p.scans.scan_00.data = u.Param()
p.scans.scan_00.data.name = 'Hdf5Loader'
p.scans.scan_00.data.orientation = 0
In addition, we assume that no meta information has been provided from the file and we need to set the photon energy, detector distance and detector pixel size directly
p.scans.scan_00.data.energy = 0.810
p.scans.scan_00.data.distance = 0.072
p.scans.scan_00.data.psize = 22e-6
which unfortunately results does not give the correct reconstruction
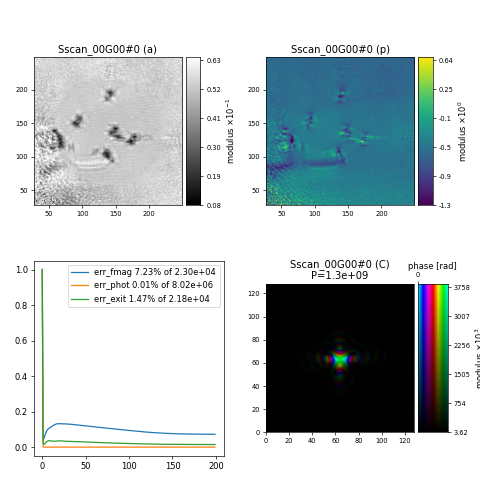
Challenge
One of the physical parameters energy, distance and psize is wrong and the data has deliberately been flipped which means that orientation needs to be adjusted. Can you fix the issues and get the reconstruction to look as expected? Hint: compare with the previous tutorial.
import ptypy, os
import ptypy.utils as u
# This will import the HDF5Loader class
ptypy.load_ptyscan_module("hdf5_loader")
# This will import the GPU engines
ptypy.load_gpu_engines("cupy")
# Root directory of tutorial data
tutorial_data_home = "../../data/"
# Dataset for this tutorial
dataset = "small_data/dls_i08_nanogold_issue.h5"
# Absolute path to HDF5 file with raw data
path_to_data = os.path.join(tutorial_data_home, dataset)
# Create parameter tree
p = u.Param()
# Set verbose level to interactive
p.verbose_level = "interactive"
# Set io settings (no files saved)
p.io = u.Param()
p.io.rfile = None
p.io.autosave = u.Param(active=False)
p.io.interaction = u.Param(active=False)
# Live-plotting during the reconstruction
p.io.autoplot = u.Param()
p.io.autoplot.active=True
p.io.autoplot.threaded = False
p.io.autoplot.layout = "jupyter"
p.io.autoplot.interval = 10
# Define the scan model
p.scans = u.Param()
p.scans.scan_00 = u.Param()
p.scans.scan_00.name = 'Full'
# Initial illumination (based on simulated optics)
p.scans.scan_00.illumination = u.Param()
p.scans.scan_00.illumination.model = None
p.scans.scan_00.illumination.photons = None
p.scans.scan_00.illumination.aperture = u.Param()
p.scans.scan_00.illumination.aperture.form = "circ"
p.scans.scan_00.illumination.aperture.size = 333e-6
p.scans.scan_00.illumination.propagation = u.Param()
p.scans.scan_00.illumination.propagation.focussed = 13.725e-3
p.scans.scan_00.illumination.propagation.parallel = 45e-6
# Data loader
p.scans.scan_00.data = u.Param()
p.scans.scan_00.data.name = 'Hdf5Loader'
p.scans.scan_00.data.orientation = 0
# Read diffraction data
p.scans.scan_00.data.intensities = u.Param()
p.scans.scan_00.data.intensities.file = path_to_data
p.scans.scan_00.data.intensities.key = "data"
# Read positions data
p.scans.scan_00.data.positions = u.Param()
p.scans.scan_00.data.positions.file = path_to_data
p.scans.scan_00.data.positions.slow_key = "posy_mm"
p.scans.scan_00.data.positions.slow_multiplier = 1e-3
p.scans.scan_00.data.positions.fast_key = "posx_mm"
p.scans.scan_00.data.positions.fast_multiplier = 1e-3
# Provide meta information
p.scans.scan_00.data.energy = 0.810
p.scans.scan_00.data.distance = 0.072
p.scans.scan_00.data.psize = 22e-6
# Define reconstruction engine (using DM)
p.engines = u.Param()
p.engines.engine = u.Param()
p.engines.engine.name = "DM_cupy"
p.engines.engine.numiter = 200
p.engines.engine.numiter_contiguous = 1
p.engines.engine.alpha = 0.9
p.engines.engine.probe_support = None
p.engines.engine.probe_update_start = 0
# Run reconstruction
P = ptypy.core.Ptycho(p,level=5)


