Chaining Multiple Engines#
Learning how to chain multiple engines and running them back to back.
A common approach in ptychography is to use one algorithm (e.g. DM or ePIE) as a starting point and another algorithm (e.g. ML) for refinement. In PtyPy, this can be achieved by simply specifying multiple engines in the parameter tree. Continuing with the same data set from the previous tutorial on Working With Electron Data we can now add “ML_pycuda” as a second engine to run after “EPIE_pycuda”
p.engines = u.Param()
p.engines.engine1 = u.Param()
p.engines.engine1.name = "EPIE_cupy"
p.engines.engine1.numiter = 10
p.engines.engine1.numiter_contiguous = 1
p.engines.engine1.alpha = 0.9
p.engines.engine1.beta = 0.1
p.engines.engine1.probe_update_start = 0
p.engines.engine1.object_norm_is_global = True
p.engines.engine2 = u.Param()
p.engines.engine2.name = "ML_cupy"
p.engines.engine2.numiter = 190
p.engines.engine2.numiter_contiguous = 1
p.engines.engine2.reg_del2 = True
p.engines.engine2.reg_del2_amplitude = 0.01
p.engines.engine2.scale_precond = True
p.engines.engine2.probe_support = None
p.engines.engine2.probe_update_start = 0
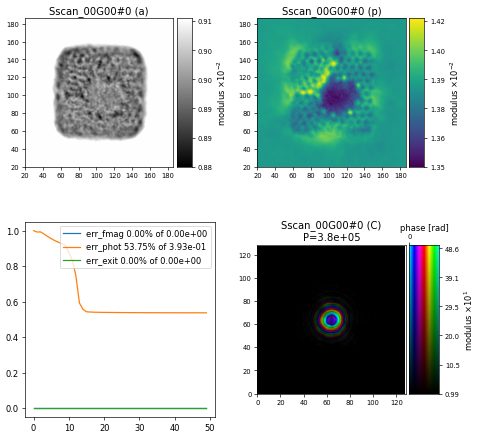
with \(40\) iterations used for the refinement and default parameters for the ML pre-conditioner and regulariser resulting in this final reconstruction
Challenge
Modify the ePIE/ML engine parameters and observe if/how it improves the quality or convergence of the reconstruction.
import ptypy, os
import ptypy.utils as u
# This will import the HDF5Loader class
ptypy.load_ptyscan_module("hdf5_loader")
# This will import the GPU engines
ptypy.load_gpu_engines("cupy")
# Root directory of tutorial data
tutorial_data_home = "../../data/"
# Dataset for this tutorial
dataset = "small_data/dls_epsic_graphene_small.h5"
# Absolute path to HDF5 file with raw data
path_to_data = os.path.join(tutorial_data_home, dataset)
# Create parameter tree
p = u.Param()
# Set verbose level to interactive
p.verbose_level = "interactive"
# Set home path and io settings (no files saved)
p.io = u.Param()
p.io.rfile = None
p.io.autosave = u.Param(active=False)
p.io.interaction = u.Param(active=False)
# Live-plotting during the reconstruction
p.io.autoplot = u.Param()
p.io.autoplot.active=True
p.io.autoplot.threaded = False
p.io.autoplot.layout = "jupyter"
p.io.autoplot.interval = 10
# Define the scan model
p.scans = u.Param()
p.scans.scan_00 = u.Param()
p.scans.scan_00.name = 'GradFull'
# Initial illumination (based on simulated optics)
p.scans.scan_00.illumination = u.Param()
p.scans.scan_00.illumination.model = None
p.scans.scan_00.illumination.photons = None
p.scans.scan_00.illumination.aperture = u.Param()
p.scans.scan_00.illumination.aperture.form = "circ"
p.scans.scan_00.illumination.aperture.size = 0.011
p.scans.scan_00.illumination.propagation = u.Param()
p.scans.scan_00.illumination.propagation.focussed = 0.1732
p.scans.scan_00.illumination.propagation.parallel = -1.0e-8
# Data loader
p.scans.scan_00.data = u.Param()
p.scans.scan_00.data.name = 'Hdf5Loader'
p.scans.scan_00.data.orientation = 0
# Read diffraction data
p.scans.scan_00.data.intensities = u.Param()
p.scans.scan_00.data.intensities.file = path_to_data
p.scans.scan_00.data.intensities.key = "data"
# Read positions data
p.scans.scan_00.data.positions = u.Param()
p.scans.scan_00.data.positions.file = path_to_data
p.scans.scan_00.data.positions.slow_key = "posy_m"
p.scans.scan_00.data.positions.slow_multiplier = 1
p.scans.scan_00.data.positions.fast_key = "posx_m"
p.scans.scan_00.data.positions.fast_multiplier = 1
# Read meta data: electron energy
p.scans.scan_00.data.electron_data = True
p.scans.scan_00.data.recorded_energy = u.Param()
p.scans.scan_00.data.recorded_energy.file = path_to_data
p.scans.scan_00.data.recorded_energy.key = "energy_kev"
p.scans.scan_00.data.recorded_energy.multiplier = 1
# Read meta data: detector distance
p.scans.scan_00.data.recorded_distance = u.Param()
p.scans.scan_00.data.recorded_distance.file = path_to_data
p.scans.scan_00.data.recorded_distance.key = "det_distance_m"
p.scans.scan_00.data.recorded_distance.multiplier = 1
# Read meta data: detector pixelsize
p.scans.scan_00.data.recorded_psize = u.Param()
p.scans.scan_00.data.recorded_psize.file = path_to_data
p.scans.scan_00.data.recorded_psize.key = "det_pixelsize_um"
p.scans.scan_00.data.recorded_psize.multiplier = 1e-6
# Read detector mask
p.scans.scan_00.data.mask = u.Param()
p.scans.scan_00.data.mask.file = path_to_data
p.scans.scan_00.data.mask.key = "mask"
# Determine diffraction center from the data
p.scans.scan_00.data.auto_center = True
# Define reconstruction engine (using ePIE)
p.engines = u.Param()
p.engines.engine1 = u.Param()
p.engines.engine1.name = "EPIE_cupy"
p.engines.engine1.numiter = 10
p.engines.engine1.numiter_contiguous = 1
p.engines.engine1.alpha = 0.9
p.engines.engine1.beta = 0.1
p.engines.engine1.probe_update_start = 0
p.engines.engine1.object_norm_is_global = True
p.engines.engine2 = u.Param()
p.engines.engine2.name = "ML_cupy"
p.engines.engine2.numiter = 40
p.engines.engine2.numiter_contiguous = 1
p.engines.engine2.reg_del2 = True
p.engines.engine2.reg_del2_amplitude = 0.01
p.engines.engine2.scale_precond = True
p.engines.engine2.probe_support = None
p.engines.engine2.probe_update_start = 0
# Run reconstruction
P = ptypy.core.Ptycho(p,level=5)


