Missing Detector Frames#
Learning how to deal with missing detector frames and how bad they can affect the reconstruction.
In general, ptychography is a quite robust phase retrieval technique, but in some cases, small inconsistencies in the data can have a big negative impact on the reconstruction. In this example, we can see what happens if we have a few missing data frames present in our data set.
The data#
Data file |
Type |
Download |
Courtesy of |
Refernce |
|---|---|---|---|---|
i08-1-5778_cropped.h5 |
Raw data |
Majid Kazemian |
We are again revisting nanogold data from I08-1, similar to the data used in Working With Large Data but this time collected with a raster grid scan (instead of a spiral scan). The raw data is located in the HDF5/Nexus file "dls_i08_nanogold_raster/i08-1-5778_cropped.h5" with the following relevant entries
* data shape = (30, 30, 1024, 1024)
* posx shape = (30, 30)
* posy shape = (30, 30)
As a first step, it often makes sense to have a look at the STXM image, which is essentially the integration of every diffraction frame. For grid scans this immediately gives us a 2D image
import h5py, os
tutorial_data_home = "../../data/"
dataset = "dls_i08_nanogold_raster/i08-1-5778_cropped.h5"
path_to_data = os.path.join(tutorial_data_home, dataset)
with h5py.File(path_to_data) as f:
stxm_image = f["datasum"][:].squeeze()
that we can plot using matplotlib
import matplotlib.pyplot as plt
plt.figure(figsize=(5,5))
plt.imshow(stxm_image, cmap="gray")
plt.show()
and observe that there is a missing frame as seen by a black dot in the image below
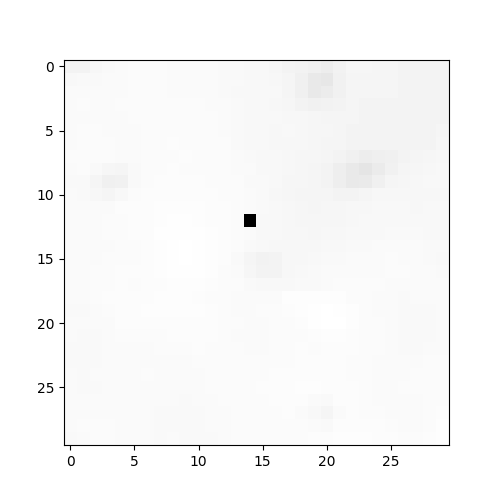
Loading the data#
We can load our data the usual way, reading the intensities
p.scans.scan_00.data.intensities = u.Param()
p.scans.scan_00.data.intensities.file = path_to_data
p.scans.scan_00.data.intensities.key = "data"
and scan positions
p.scans.scan_00.data.positions = u.Param()
p.scans.scan_00.data.positions.file = path_to_data
p.scans.scan_00.data.positions.slow_key = "posy"
p.scans.scan_00.data.positions.slow_multiplier = 1e-3
p.scans.scan_00.data.positions.fast_key = "posx"
p.scans.scan_00.data.positions.fast_multiplier = 1e-3
p.scans.scan_00.data.positions.bounding_box = u.Param()
p.scans.scan_00.data.positions.bounding_box.fast_axis_bounds = [5,25]
p.scans.scan_00.data.positions.bounding_box.slow_axis_bounds = [5,25]
using the bounding_box to select a sub-region just to speed up the loading and reconstruction time for purpose of the example.
Reconstruction engine#
Similar to the previous I08-1 nanogold example (Working With Large Data) we use DM with the following set of parameters
p.engines = u.Param()
p.engines.engine = u.Param()
p.engines.engine.name = "DM_pycuda"
p.engines.engine.numiter = 500
p.engines.engine.numiter_contiguous = 10
p.engines.engine.probe_support = None
p.engines.engine.probe_update_start = 0
p.engines.engine.probe_fourier_support = None
p.engines.engine.record_local_error = False
p.engines.engine.alpha = 0.95
p.engines.engine.fourier_power_bound = 0.25
p.engines.engine.overlap_converge_factor = 0.001
p.engines.engine.overlap_max_iterations = 20
p.engines.engine.update_object_first = False
p.engines.engine.obj_smooth_std = 20
p.engines.engine.object_inertia = 0.001
p.engines.engine.probe_inertia = 0.001
p.engines.engine.clip_object = [0,1]
which will result in the following reconstruction after \(500\) iterations
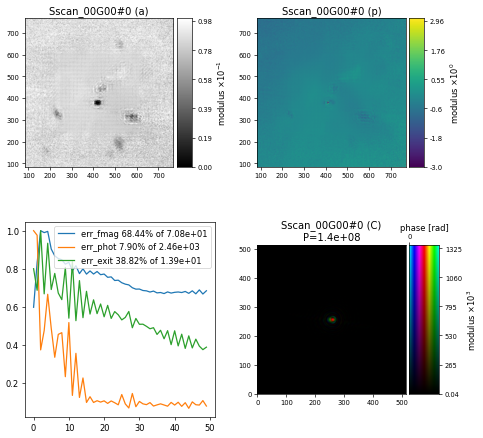
which clearly shows that something went a bit wrong here, likely because of the errant data frames.
Ignore missing frames#
To fix this issue, we can simply tell the "Hdf5Loader" to ignore those missing frames by using a framefilter for which we can use the STXM image that we already looked at above
p.scans.scan_00.data.framefilter = u.Param()
p.scans.scan_00.data.framefilter.file = path_to_data
p.scans.scan_00.data.framefilter.key = "entry/axis_sum/sum"
With this small change, the reconstruction after \(500\) iterations of DM now looks much better
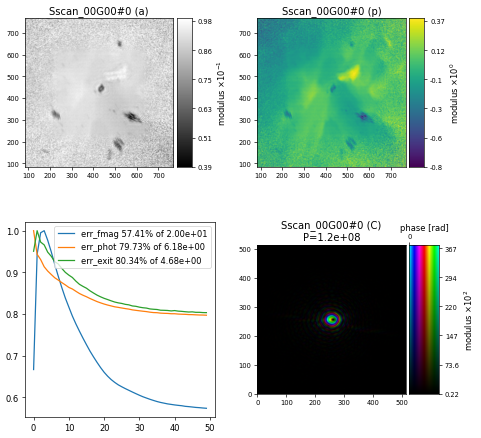
Challenge
Compare reconstructions without and with using the framefilter to exclude the missing detector frame.
import ptypy, os
import ptypy.utils as u
# This will import the HDF5Loader class
ptypy.load_ptyscan_module("hdf5_loader")
# This will import the GPU engines
ptypy.load_gpu_engines("cupy")
# Root directory of tutorial data
tutorial_data_home = "../../data/"
# Dataset for this tutorial
dataset = "dls_i08_nanogold_raster/i08-1-5778_cropped.h5"
# Absolute path to HDF5 file with raw data
path_to_data = os.path.join(tutorial_data_home, dataset)
# Create parameter tree
p = u.Param()
# Set verbose level to info
p.verbose_level = "interactive"
# Scan label
p.run = "dls_i08_nanogold"
# Data loading and processing should
# happen in chunks of this size
p.frames_per_block = 100
# Set io settings (no files saved)
p.io = u.Param()
p.io.rfile = None
p.io.autosave = u.Param(active=False)
p.io.interaction = u.Param(active=False)
# Live-plotting during the reconstruction
p.io.autoplot = u.Param()
p.io.autoplot.active=True
p.io.autoplot.threaded = False
p.io.autoplot.layout = "jupyter"
p.io.autoplot.interval = 10
# Define the scan model
p.scans = u.Param()
p.scans.scan_00 = u.Param()
p.scans.scan_00.name = 'BlockFull'
# Initial illumination (based on simulated optics)
p.scans.scan_00.illumination = u.Param()
p.scans.scan_00.illumination.model = None
p.scans.scan_00.illumination.photons = None
p.scans.scan_00.illumination.aperture = u.Param()
p.scans.scan_00.illumination.aperture.form = "circ"
p.scans.scan_00.illumination.aperture.size = 333e-6
p.scans.scan_00.illumination.propagation = u.Param()
p.scans.scan_00.illumination.propagation.focussed = 13.725e-3
p.scans.scan_00.illumination.propagation.parallel = 30e-6
p.scans.scan_00.illumination.propagation.antialiasing = 1
p.scans.scan_00.illumination.diversity = u.Param()
p.scans.scan_00.illumination.diversity.power = 0.1
p.scans.scan_00.illumination.diversity.noise = [0.5,1.0]
# Initial object
p.scans.scan_00.sample = u.Param()
p.scans.scan_00.sample.model = None
p.scans.scan_00.sample.diversity = None
p.scans.scan_00.sample.process = None
# Coherence parameters (modes)
p.scans.scan_00.coherence = u.Param()
p.scans.scan_00.coherence.num_probe_modes = 1
p.scans.scan_00.coherence.num_object_modes = 1
# Data loader
p.scans.scan_00.data = u.Param()
p.scans.scan_00.data.name = 'Hdf5LoaderFast'
p.scans.scan_00.data.orientation = 2
p.scans.scan_00.data.intensities = u.Param()
p.scans.scan_00.data.intensities.file = path_to_data
p.scans.scan_00.data.intensities.key = "data"
p.scans.scan_00.data.positions = u.Param()
p.scans.scan_00.data.positions.file = path_to_data
p.scans.scan_00.data.positions.slow_key = "posy"
p.scans.scan_00.data.positions.slow_multiplier = 1e-3
p.scans.scan_00.data.positions.fast_key = "posx"
p.scans.scan_00.data.positions.fast_multiplier = 1e-3
p.scans.scan_00.data.positions.bounding_box = u.Param()
p.scans.scan_00.data.positions.bounding_box.fast_axis_bounds = [5,25]
p.scans.scan_00.data.positions.bounding_box.slow_axis_bounds = [5,25]
p.scans.scan_00.data.recorded_energy = u.Param()
p.scans.scan_00.data.recorded_energy.file = path_to_data
p.scans.scan_00.data.recorded_energy.key = "energy"
p.scans.scan_00.data.recorded_energy.multiplier = 1e-3
p.scans.scan_00.data.darkfield = u.Param()
p.scans.scan_00.data.darkfield.file = path_to_data
p.scans.scan_00.data.darkfield.key = "dark"
# p.scans.scan_00.data.framefilter = u.Param()
# p.scans.scan_00.data.framefilter.file = path_to_data
# p.scans.scan_00.data.framefilter.key = "datasum"
p.scans.scan_00.data.energy = 0.710
p.scans.scan_00.data.distance = 0.072
p.scans.scan_00.data.psize = 11e-6
p.scans.scan_00.data.shape = (1024,1024)
p.scans.scan_00.data.rebin = 2
# Reconstruct using GPU-accelerated DM
p.engines = u.Param()
p.engines.engine = u.Param()
p.engines.engine.name = "DM_cupy"
p.engines.engine.numiter = 500
p.engines.engine.numiter_contiguous = 10
p.engines.engine.probe_support = None
p.engines.engine.probe_update_start = 0
p.engines.engine.probe_fourier_support = None
p.engines.engine.record_local_error = False
p.engines.engine.alpha = 0.95
p.engines.engine.fourier_power_bound = 0.25
p.engines.engine.overlap_converge_factor = 0.001
p.engines.engine.overlap_max_iterations = 20
p.engines.engine.update_object_first = False
p.engines.engine.obj_smooth_std = 20
p.engines.engine.object_inertia = 0.001
p.engines.engine.probe_inertia = 0.001
p.engines.engine.clip_object = [0,1]
# Run reconstruction
P = ptypy.core.Ptycho(p,level=5)


