Nearfield Ptychography#
Learning how to reconstruct a nearfield ptychography dataset with PtyPy.
All examples so far were based on farfield ptychography, but some users might be interested to reconstruct data from a nearfield ptychography experiment. In this example, we are going to use data that was published in this article about Phase-Vortex Removal for Quantitative X-Ray Nanotomography with Near-Field Ptychography and has kindly been provided by the authors to be used in this tutorial.
The data#
Thanks to Pierre Thibault for sharing this data set for the purpose of this workshop.
The sample used for this nearfield ptychography experiment is made of Al and Ni particles inside a glass capillary and the data was collected at the ID16 instrument at the ESRF. The raw data is saved in an HDF5 file with the following entries
* data shape = (17, 2048, 2048)
* posx shape = (17,)
* posy shape = (17,)
showing that we have \(17\) nearfield diffraction images of size \(2048x2048\).
Loading the data#
We can use the Hdf5Loader
p.scans.scan_00.data = u.Param()
p.scans.scan_00.data.name = 'Hdf5Loader'
p.scans.scan_00.data.orientation = 3
to load the intensities
p.scans.scan_00.data.intensities = u.Param()
p.scans.scan_00.data.intensities.file = path_to_data
p.scans.scan_00.data.intensities.key = 'data'
and scan positions
p.scans.scan_00.data.positions = u.Param()
p.scans.scan_00.data.positions.file = path_to_data
p.scans.scan_00.data.positions.slow_key = 'posy'
p.scans.scan_00.data.positions.slow_multiplier = 1
p.scans.scan_00.data.positions.fast_key = 'posx'
p.scans.scan_00.data.positions.fast_multiplier = 1
We also need to specify some meta information
p.scans.scan_00.data.auto_center = False
p.scans.scan_00.data.psize = 50.705e-9
p.scans.scan_00.data.distance = 0.019
p.scans.scan_00.data.energy = 17.05
Note
Nearfield ptychography experiments are usually performed in cone-beam geometry and therefore the estimate of the sample-to-detector distance needs to be adjusted such that it can be used within parallel geometry models, see this article for more information.
The scan model#
Since we are only a few frames, it is reasonable to use the "Full" scan model, but we need to switch propagation to nearfield
p.scans = u.Param()
p.scans.scan_00 = u.Param()
p.scans.scan_00.name = 'Full'
p.scans.scan_00.propagation = "nearfield"
Initial probe#
For a nearfield ptychography reconstruction, the best starting guess for the illumination would be a flatfield (beam on but without sample) back-propagated into the sample plane. If there is no flatfield available, it is also reasonable to simply average over all raw data frames and back-propagate the result into the sample plane. This can be achieved by choosing the "stxm" option the illumination.model
p.scans.scan_00.illumination = u.Param()
p.scans.scan_00.illumination.model = "stxm"
p.scans.scan_00.illumination.aperture = None
Reconstruction engine#
For the reconstruction, we have chosen a combination of DM followed by ML refinement with the following set of parameters
p.engines = u.Param()
p.engines.DM = u.Param()
p.engines.DM.name = 'DM_pycuda'
p.engines.DM.numiter = 500
p.engines.DM.numiter_contiguous = 10
p.engines.DM.probe_support = None
p.engines.DM.clip_object = (.2, 1.1)
p.engines.DM.fourier_power_bound = 0.0
p.engines.DM.probe_update_start = 0
p.engines.ML = u.Param()
p.engines.ML.name = 'ML_pycuda'
p.engines.ML.numiter = 500
p.engines.ML.numiter_contiguous = 10
p.engines.ML.ML_type = 'Gaussian'
p.engines.ML.floating_intensities = False
p.engines.ML.probe_support = None
p.engines.ML.reg_del2 = True
p.engines.ML.reg_del2_amplitude = 0.005
p.engines.ML.scale_precond = False
p.engines.ML.probe_update_start = 0
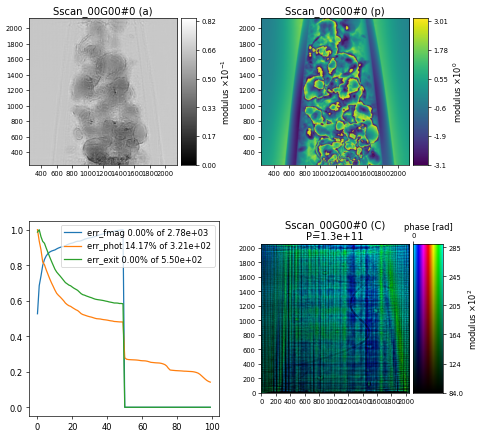
resulting in the following reconstruction of the AlNi sample
import ptypy, h5py, os
import ptypy.utils as u
# This will import the HDF5Loader class
ptypy.load_ptyscan_module("hdf5_loader")
# This will import the GPU engines
ptypy.load_gpu_engines("cupy")
# Root directory of tutorial data
tutorial_data_home = "../../data/"
# Path to HDF5 file with raw data
dataset = "esrf_id16_AlNi_nearfield/S00084_data_bin1_newpos_2048x2048.h5"
# Absolute path to HDF5 file with raw data
path_to_data = os.path.join(tutorial_data_home, dataset)
# Create parameter tree
p = u.Param()
# Set verbose level to interactive
p.verbose_level = "interactive"
# Scan label
p.run = "esrf_id16_AlNi_nearfield"
# Set io settings (no files saved)
p.io = u.Param()
p.io.rfile = None
p.io.autosave = u.Param(active=False)
p.io.interaction = u.Param(active=False)
# Live-plotting during the reconstruction
p.io.autoplot = u.Param()
p.io.autoplot.active=True
p.io.autoplot.threaded = False
p.io.autoplot.layout = "jupyter"
p.io.autoplot.interval = 10
# Define the scan model
p.scans = u.Param()
p.scans.scan_00 = u.Param()
p.scans.scan_00.name = 'Full'
# Switch propagation to nearfield
p.scans.scan_00.propagation = "nearfield"
# Initial illumination (based on flatfield)
p.scans.scan_00.illumination = u.Param()
p.scans.scan_00.illumination.model = "stxm"
p.scans.scan_00.illumination.aperture = None
# Initial object
p.scans.scan_00.sample = u.Param()
p.scans.scan_00.sample.model = None
p.scans.scan_00.sample.diversity = None
p.scans.scan_00.sample.process = None
# Coherence parameters (modes)
p.scans.scan_00.coherence = u.Param()
p.scans.scan_00.coherence.num_probe_modes = 1
p.scans.scan_00.coherence.num_object_modes = 1
# Data loader
p.scans.scan_00.data = u.Param()
p.scans.scan_00.data.name = 'Hdf5Loader'
p.scans.scan_00.data.orientation = 3
p.scans.scan_00.data.intensities = u.Param()
p.scans.scan_00.data.intensities.file = path_to_data
p.scans.scan_00.data.intensities.key = 'data'
p.scans.scan_00.data.positions = u.Param()
p.scans.scan_00.data.positions.file = path_to_data
p.scans.scan_00.data.positions.slow_key = 'posy'
p.scans.scan_00.data.positions.slow_multiplier = 1
p.scans.scan_00.data.positions.fast_key = 'posx'
p.scans.scan_00.data.positions.fast_multiplier = 1
# Read meta information
p.scans.scan_00.data.auto_center = False
p.scans.scan_00.data.psize = 50.705e-9
p.scans.scan_00.data.distance = 0.019
p.scans.scan_00.data.energy = 17.05
# Reconstruct using GPU-accelerated DM/ML engines
p.engines = u.Param()
p.engines.DM = u.Param()
p.engines.DM.name = 'DM_cupy'
p.engines.DM.numiter = 500
p.engines.DM.numiter_contiguous = 10
p.engines.DM.probe_support = None
p.engines.DM.clip_object = (.2, 1.1)
p.engines.DM.fourier_power_bound = 0.0
p.engines.DM.probe_update_start = 0
p.engines.ML = u.Param()
p.engines.ML.name = 'ML_cupy'
p.engines.ML.numiter = 500
p.engines.ML.numiter_contiguous = 10
p.engines.ML.ML_type = 'Gaussian'
p.engines.ML.floating_intensities = False
p.engines.ML.probe_support = None
p.engines.ML.reg_del2 = True
p.engines.ML.reg_del2_amplitude = 0.005
p.engines.ML.scale_precond = False
p.engines.ML.probe_update_start = 0
# Run reconstruction
P = ptypy.core.Ptycho(p,level=5)


